AMGT-TS User manual (web-based version)
Create a new project
Step 1:
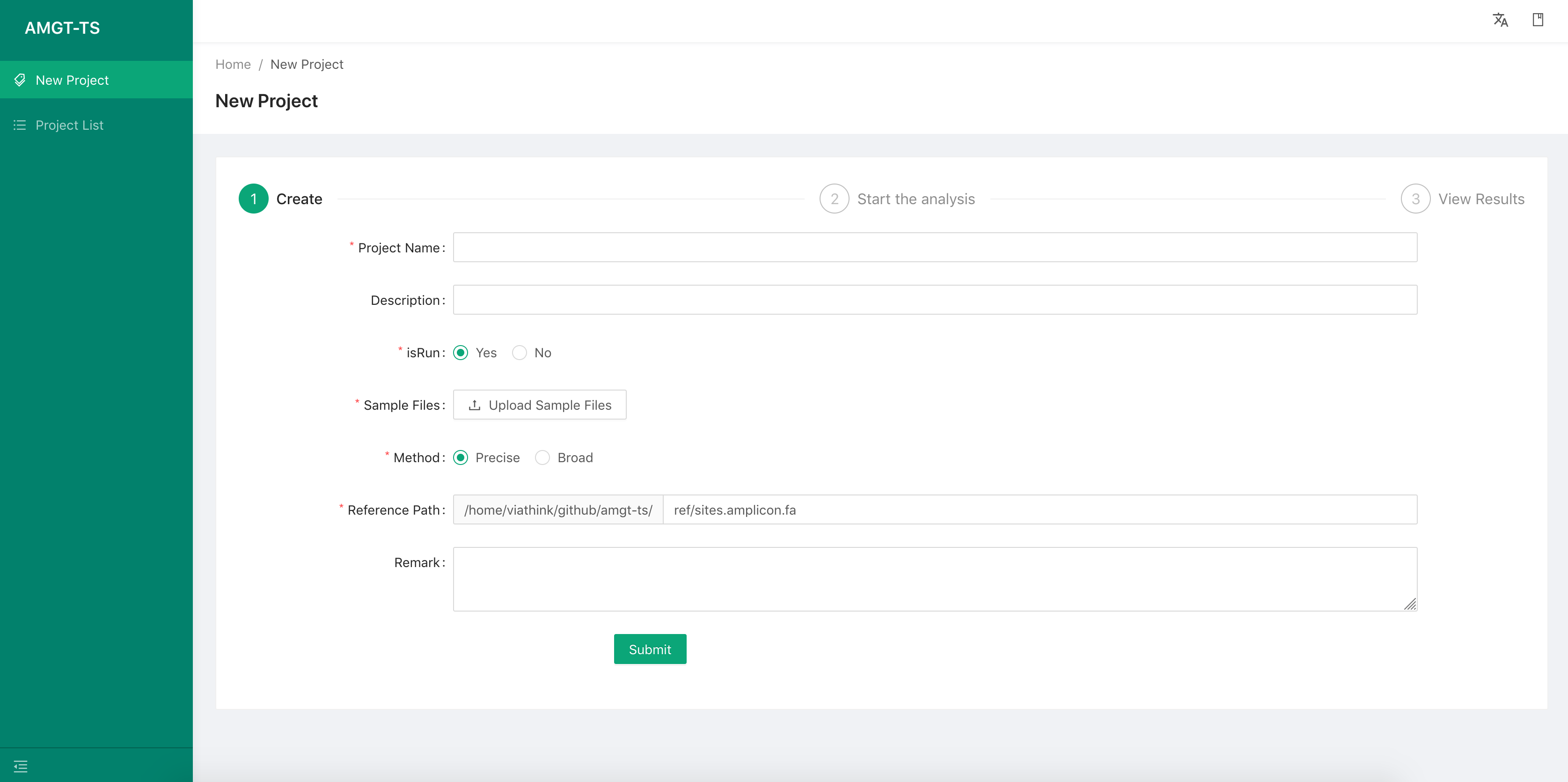
Launch this function by select the menu item “New Project” on the left, then fill the following information:
-
Project name
-
Project description
-
Upload the targeted sequencing file of each sample
we can upload more than one samples data files and there is no limit.
-
Reference files Use the default if you are not familiar with the details.
-
Remark
Then use the Next button to continue.
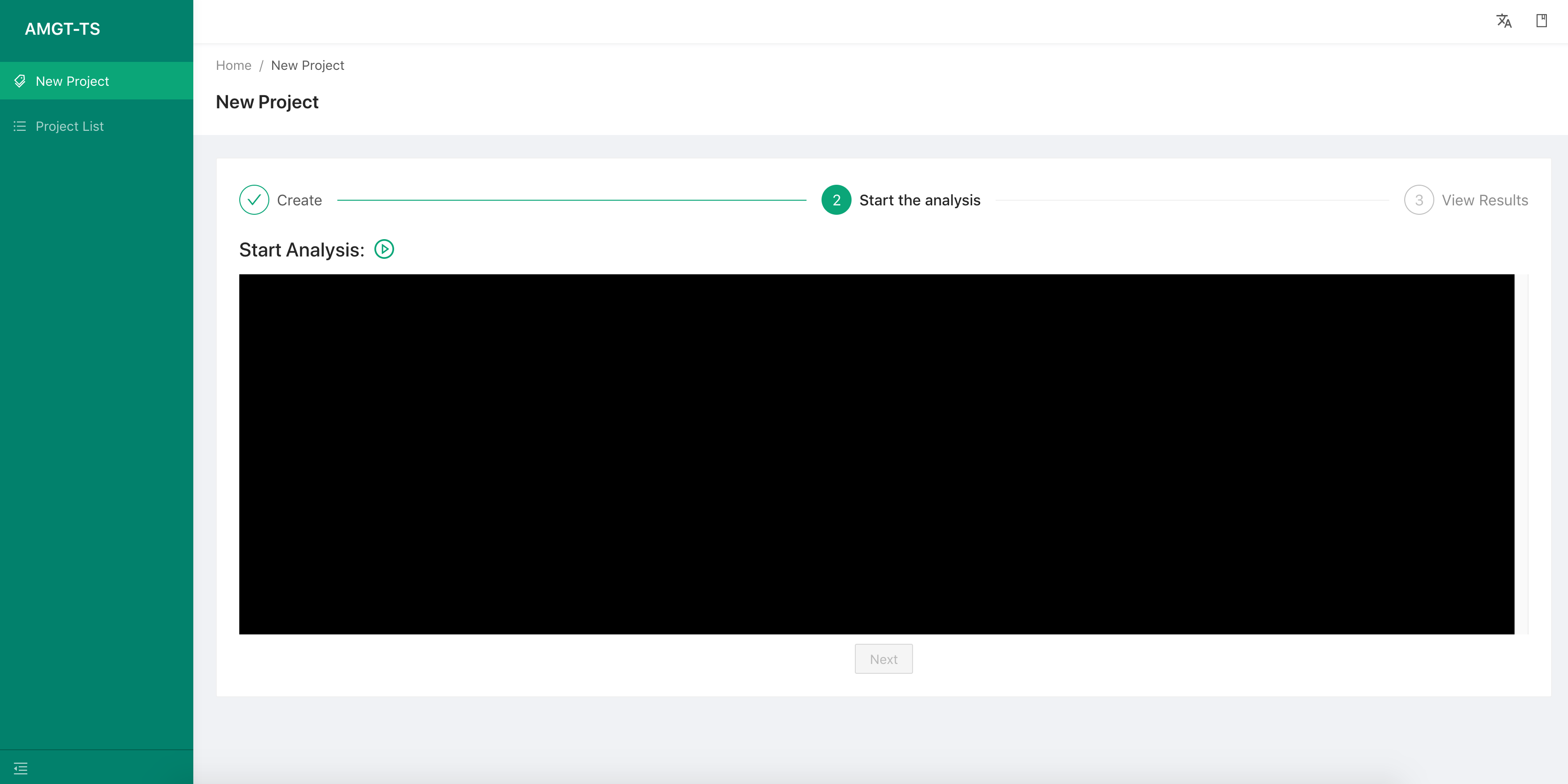
Step 2:
-
After creating the project, we use the
Startbutton to launch the AMGT-TS tool. -
Then, we use the
Nextbutton to go to the result tab page.
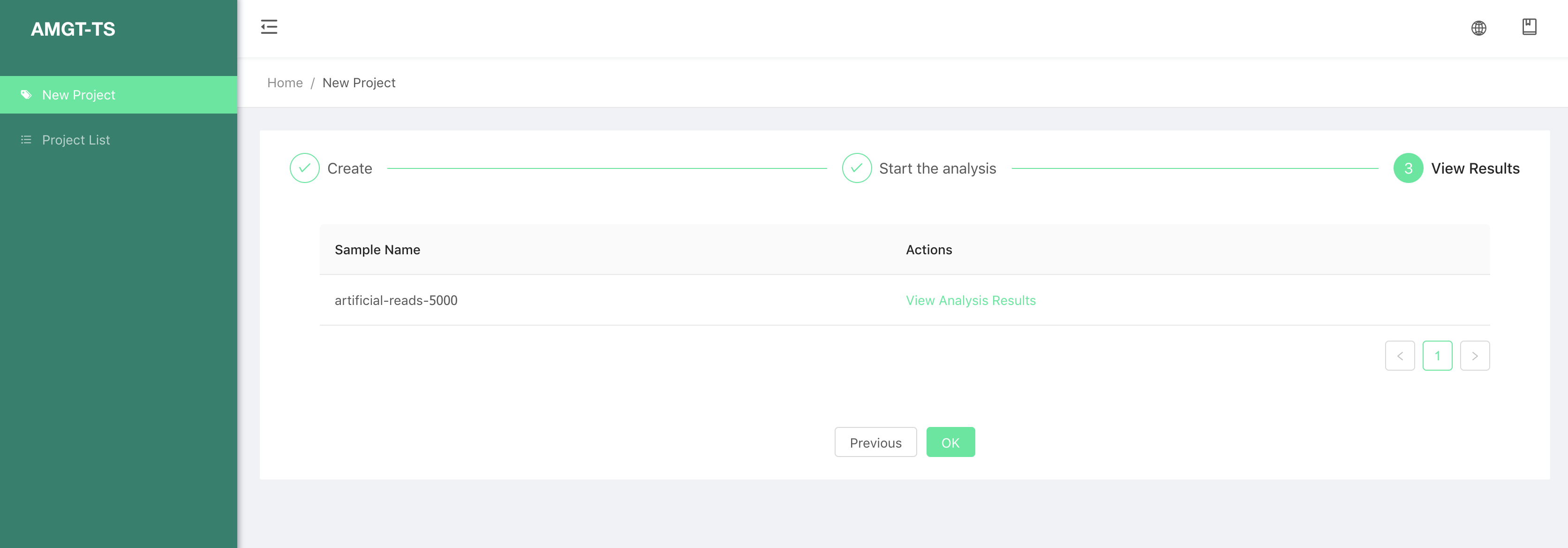
Step 3:
- This page show the analysis result of AMGT-TS tool.
We can select the View Analysis Results of a sample, to view all the loci result information of this sample.
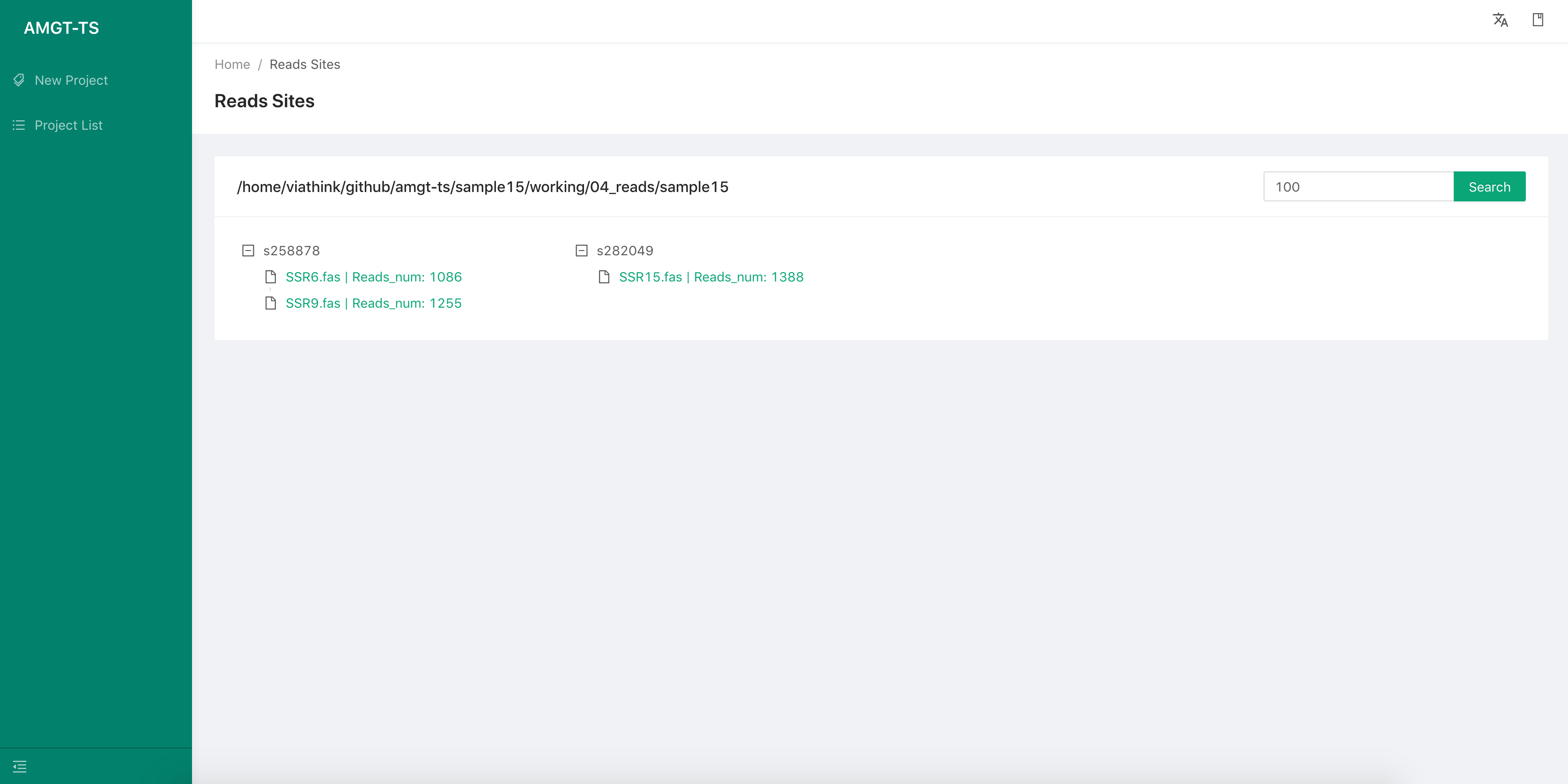
- We can show the reads alignment graph by select a locus. In the graph, the first read is the reference read of the locus,others is from the targeted sequencing data.

Project List
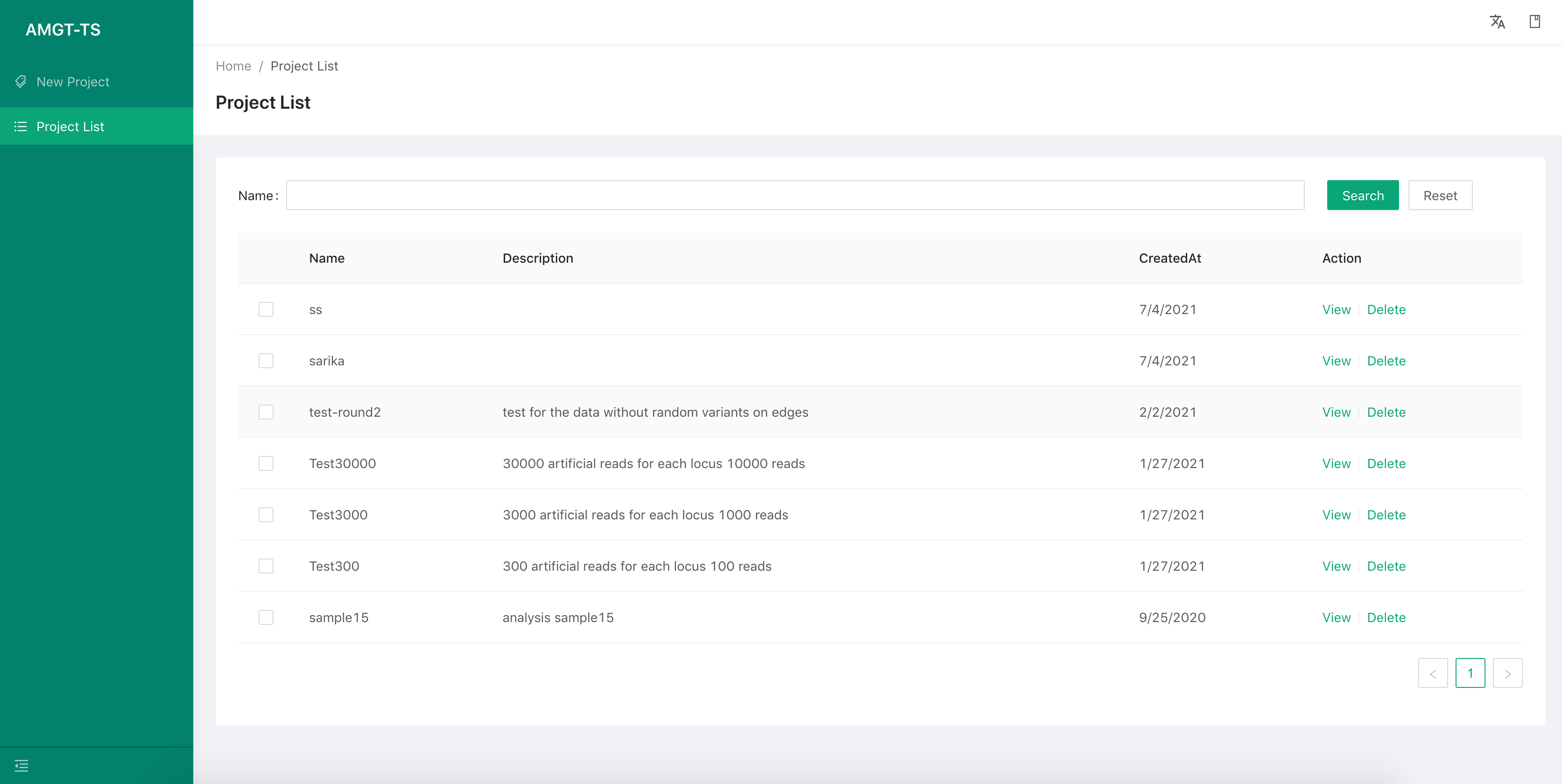
We can view all the projects information by select the second menu item on the left. By select the View button to check the details of each project. There are information of each project as following:
-
Basic information Such as project name, description, the result path, etc.
-
Samples list: Same as the step 3 of “Create a new project”.